CRISPR, Gene Drive Technology, and Hope for the Future

Contents
The following article was written by John Min and George Church.
Imagine for a moment, a world where we are able to perform genetic engineering on such large scales as to effectively engineer nature. In this world, parasites that only cause misery and suffering would not exist, only minimal pesticides and herbicides would be necessary in agriculture, and the environment would be better adapted to maximize positive interactions with all human activities while maintaining sustainability. While this may all sound like science fiction, the technology that might allow us to reach this utopia is very real, and if we develop it responsibly, this dream may well become reality.
‘Gene drive’ technology, or more specifically, CRISPR gene drives, have been heralded by the press as a potential solution for mosquito-borne diseases such as malaria, dengue, and most recently, Zika. In general, gene drive is a technology that allows scientists to bias the rate of inheritance of specific genes in wild populations of organisms. A gene is said to ‘drive’ when it is able to increase the frequency of its own inheritance higher than the expected probability of 50%. In doing so, gene drive systems exhibit unprecedented ability to directly manipulate genes on a population-wide scale in nature.
The idea to use gene drive systems to propagate engineered genes in natural systems is not new. Indeed, a proposal to construct gene drives using naturally occurring homing nucleases, genes that can specifically cut DNA and insert extra copies of itself, was published by Austin Burt in 2003 (Burt, 2013). In fact, the concept was discussed even before the earliest studies on naturally driving genetic elements — such as transposons, which are small sections of DNA that can insert extra copies of itself — over half a century ago (Serebrovskii, 1940) (Vanderplank, 1944).
However, it is only with advances in modern genome editing technology, such as CRISPR, that scientists are finally able to digitally target gene drives to any desired location in the genome. Ever since the first CRISPR gene drive design was described in a 2014 publication by Kevin Esvelt and George Church (Esvelt, et al., 2014), man-made gene drive systems have been successfully tested in three separate species, yeast, fruit fly, and mosquitoes (DiCarlo, et al., 2015) (Gantz & Bier, 2015) (Gantz, et al., 2015) .
The term ‘CRISPR’ stands for clustered regularly-interspaced short palindromic repeats and describes an adaptive immune system against viral infections originally discovered in bacteria. Nucleases, or proteins that cut DNA, in the CRISPR family are generally able to cut DNA anywhere as specified by a short stretch of RNA sequence at high precision and accuracy.
The nuclease cas9, in particular, has become a favorite among geneticists around the world since the publication of a series of high impact journal articles in late 2012 and early 2013 (Jinek, et al., 2012) (Cong, et al., 2013) (Hwang, et al., 2013). Using cas9, scientists are able to create ‘double-stranded breaks,’ or cuts in DNA, at nearly any location specified by a 20 nucleotide piece of RNA sequence.
After being cut, we can take advantage of natural DNA repair mechanisms to persuade cells to incorporate new genetic information into the break. This allows us to introduce new genes into an organism or even bar-code it at a genetic level. By using CRISPR technology, scientists are also able to insert synthesized gene drive systems into a host organism’s genome with the same high level of precision and reliability.
Potential applications for CRISPR gene drives are broad and numerous, as the technology is expected to work in any organism that reproduces sexually.
While popular media attention is chiefly focused on the elimination of mosquito-borne diseases, applications also exist in the fight against the rise of Lyme disease in the U.S. Beyond public health, gene drives can be used to eliminate invasive species from non-native habitats, such as mosquitos in Hawaii. In this case, many native Hawaiian bird species, especially the many honeycreepers, are being driven to extinction by mosquito-borne avian malaria. The removal of mosquitos in Hawaii would both save the bird populations, as well as make Hawaii even more attractive as a tropical paradise for tourists.
With such rapid expansion of gene drive technology over the past year, it is only natural for there to be some concern and fear over attempting to genetically engineer nature at such a large scale. The only way to truly address these fears is to rigorously test the spreading properties of various gene drive designs within the safety of the laboratory — something that has also been in active development over the last year.
It is also important to remember that mankind has been actively engineering the world around us since the dawn of civilization, albeit with more primitive tools. Using a mixture of breeding and mechanical tools, we have managed to transform teosinte into modern corn, created countless breeds of dogs and cats, and transformed vast stretches everything from lush forests to deserts into modern farmland.
Yet, these amazing feats are not without consequence. Most products of our breeding techniques are unable to survive independently in nature, and countless species have become extinct as the result of our agricultural expansion and eco-engineering.
It is imperative that we approach gene drives differently, with increased consideration for the consequences of our actions on both the natural world as well as ourselves. Proponents of gene drive technology would like to initiate a new research paradigm centered on collective decision making. As most members of the public will inevitably be affected by a gene drive release, it is only ethical to include the public throughout the research and decision making process of gene drive development. Furthermore, by being transparent and inviting of public criticism, researchers are able to crowd-source the “de-bugging” process, as well as minimize the risk of a gene drive release going awry.
We must come to terms with the reality that thousands of acres of habitat continue to be destroyed annually through a combination of chemical sprays, urban and agricultural expansion, and the introduction of invasive species, just to name a few. To improve up on this, I would like to echo the hopes of my mentor, Kevin Esvelt, toward the use of “more science, and fewer bulldozers for environmental engineering” in hopes of creating a more sustainable co-existence between man and nature. The recent advancements in CRISPR gene drive technology represent an important step toward this hopeful future.
About the author: John Min is a PhD. Candidate in the BBS program at Harvard Medical School co-advised by Professor George Church and Professor Kevin Esvelt at MIT Media Labs. He is currently working on creating a laboratory model for gene drive research.
References
Burt, A. (2013). Site-specific selfish genes as tools for the control and genetic engineering of naturl populations. Proceedings of the biological sciences B, 270:921-928.
Cong, L., Ann Ran, F., Cox, D., Lin, S., Barretto, R., Habib, N., . . . Zhang, F. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science, 819-823.
DiCarlo, J. E., Chavez, A., Dietz, S. L., Esvelt, K. M., & Church, G. M. (2015). RNA-guided gene drives can efficiently and reversibly bias inheritance in wild yeast. bioRxiv preprint, DOI:10.1101/013896.
Esvelt, K. M., Smidler, A. L., Catteruccia, F., & Church, G. M. (2014). Concerning RNA-guided gene drives for the alteration of wild populations. eLIFE, 1-21.
Gantz, V. M., & Bier, E. (2015). The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science, Vol. 348 442-444.
Gantz, V., Jasinskiene, N., Tatarenkova, O., fazekas, A., Macias, V. M., Bier, E., & James, A. A. (2015). Highly efficient Cas90mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. PNAS, vol.112 49.
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., . . . Joung, J. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature Biotechnology, 227-229.
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science, 816-821.
Serebrovskii, A. (1940). On the possibility of a new method for the control of insect pests. Zool.Zh.
Vanderplank, F. (1944). Experiments in crossbreeding tsetse flies, Gossina species. Nature, vol.144 607-608.
About the Future of Life Institute
The Future of Life Institute (FLI) is a global think tank with a team of 20+ full-time staff operating across the US and Europe. FLI has been working to steer the development of transformative technologies towards benefitting life and away from extreme large-scale risks since its founding in 2014. Find out more about our mission or explore our work.
Related content
Other posts about Biotech, Recent News
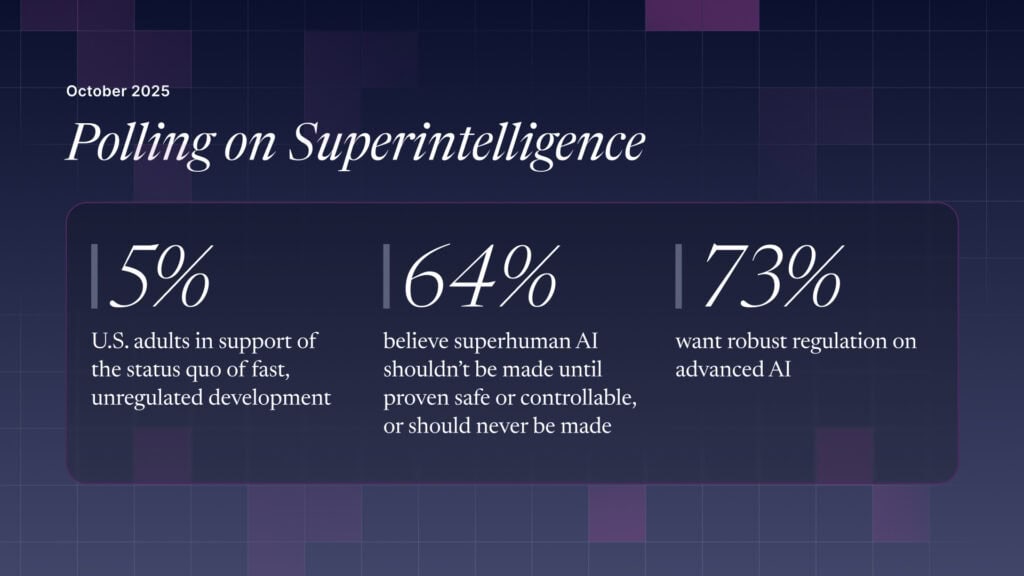
The U.S. Public Wants Regulation (or Prohibition) of Expert‑Level and Superhuman AI

Poll Shows Broad Popularity of CA SB1047 to Regulate AI